Neurons Encoding Catalytic Serines: A Case Study on Interpretability in Protein Models
Nithin Parsan
Co-Founder
Introduction
Mechanistic interpretability has proven valuable for understanding large language models, but can these techniques help us understand biological language models? In this post, we demonstrate a proof-of-concept applying sparse probing techniques to ESM-2, a protein language model trained on 65M protein sequences. We show how just two neurons encode one of biology's most fundamental features: the catalytic machinery of serine protease.
Why Serine Proteases?
Serine proteases offer an ideal test case for probing in ESM-2 because they represent a clear binary property—either a sequence has a functional catalytic serine or it doesn't. This allows us to create a clean dataset with well-defined ground truth labels:
- Positive examples: Wild-type sequences with verified catalytic activity
- Negative examples: Sequences with the catalytic serine systematically mutated to all 19 other amino acids
This binary classification task lets us distinguish between neurons that merely detect serine residues and those that specifically encode catalytic serines.
Methods
Using ESM2-t6-8M-UR50D (a 6-layer model with hidden dimension 1280), we extracted post-GELU activations from feed-forward layers and aggregated them across sequence length by taking maximum activations. Following Gurnee et al., we implemented several sparse probing methods:
- Mean activation difference between classes
- Mutual information between activations and labels
- L1-regularized logistic regression
- Support Vector Machines with hinge loss
Key Findings
Our analysis revealed two distinct mechanisms working together across layers:
-
A Precise Feature Detector in Layer 1 (Neuron 1269):
- Consistently identified across all probing methods and sparsity levels
- Shows asymmetric behavior under intervention: negative scaling reduces predictions by 70.8%
- Specifically affects catalytic serines (p = 0.029) with minimal effect on non-catalytic serines
-
A Complementary Circuit in Layer 5 (Neurons 106 and 110):
- Achieves perfect F1 score (1.000) with just k=2 neurons
- Neuron 106 shows strong positive correlation with catalytic function (activation difference: 1.560)
- Neuron 110 provides regulatory counterbalance
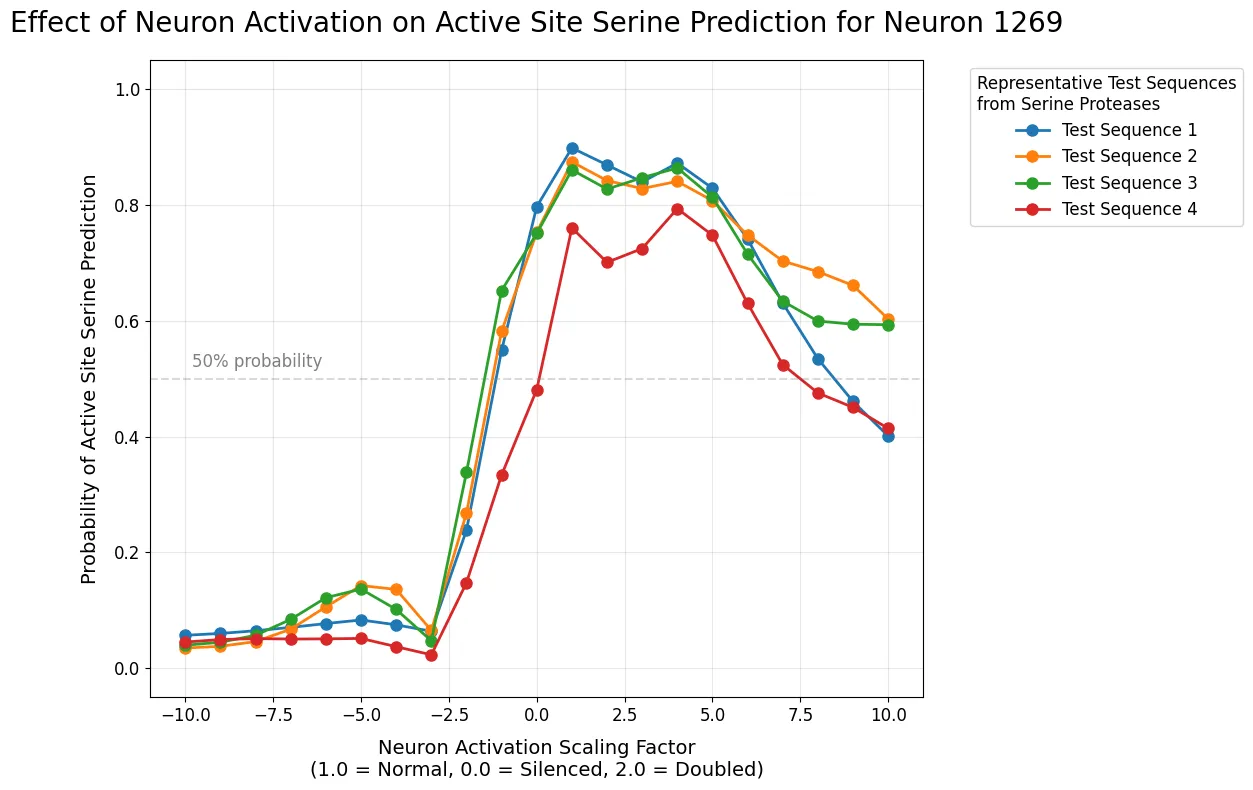
Figure 1: Effect of Neuron Activation on Active Site Serine Prediction for Neuron 1269
Graph showing how scaling Neuron 1269's activation affects probability of active site serine prediction across multiple test sequences.
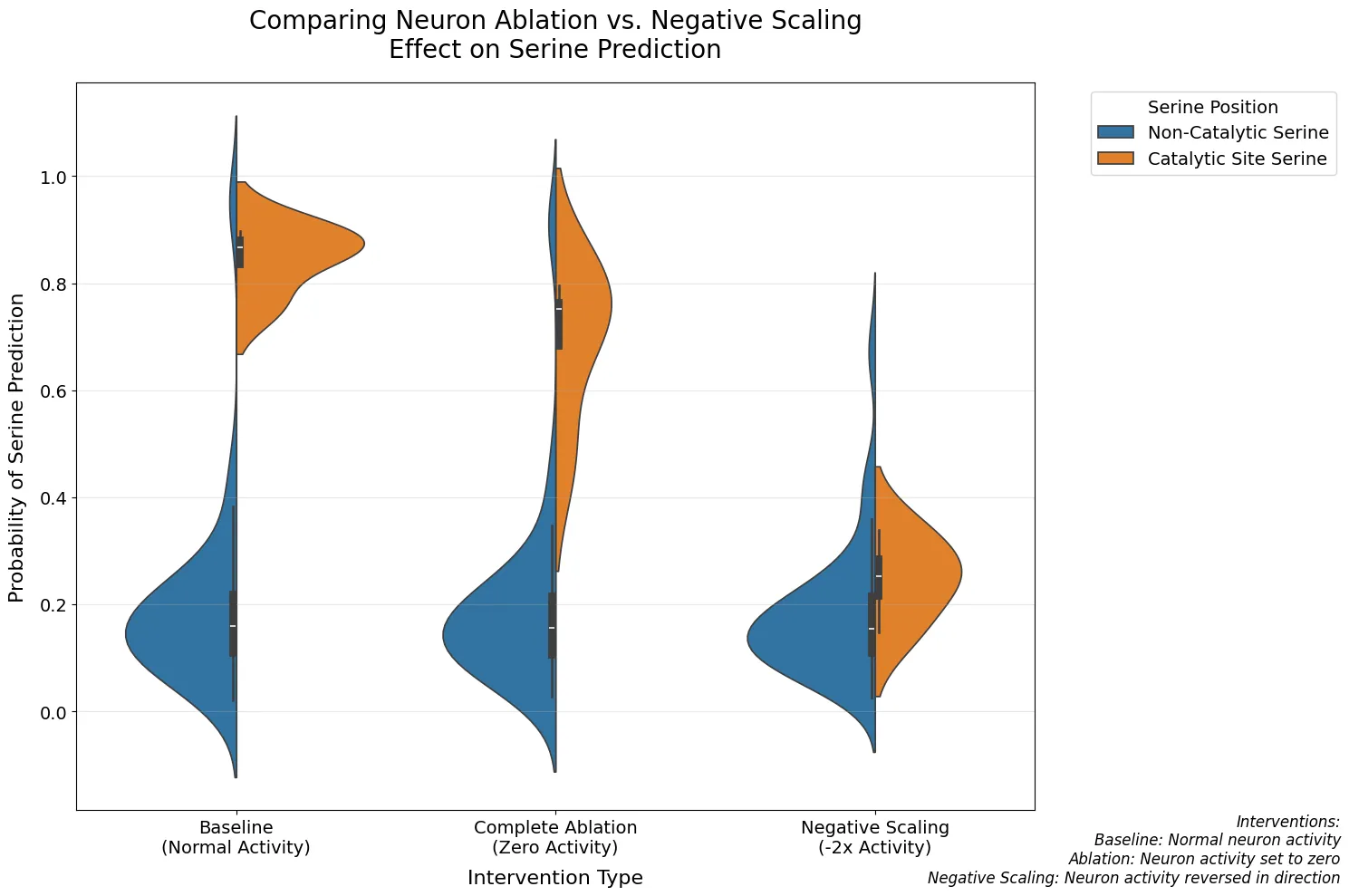
Figure 2: Comparing Neuron Ablation vs. Negative Scaling
Violin plots showing distribution of serine prediction probabilities under different intervention conditions: baseline, complete ablation, and negative scaling.
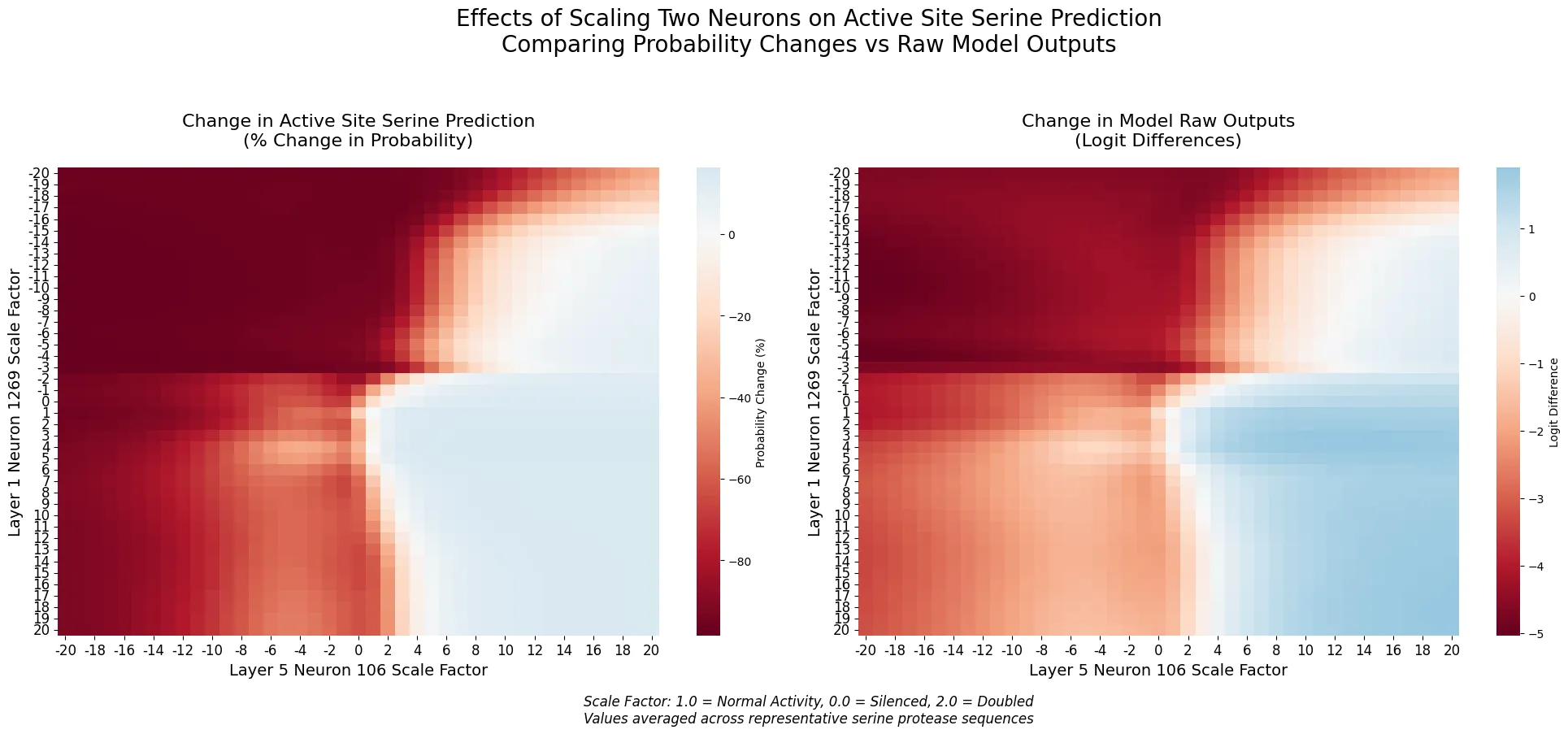
Cross-Layer Interactions
By simultaneously manipulating neurons across layers, we discovered sophisticated computational patterns:
- The early-layer neuron 1269 shows stronger modulation of layer 5 effects than vice versa
- Sharp transitions in prediction occur when both neurons shift from positive to negative scaling
- Synergistic enhancement occurs when neurons are scaled in compatible directions
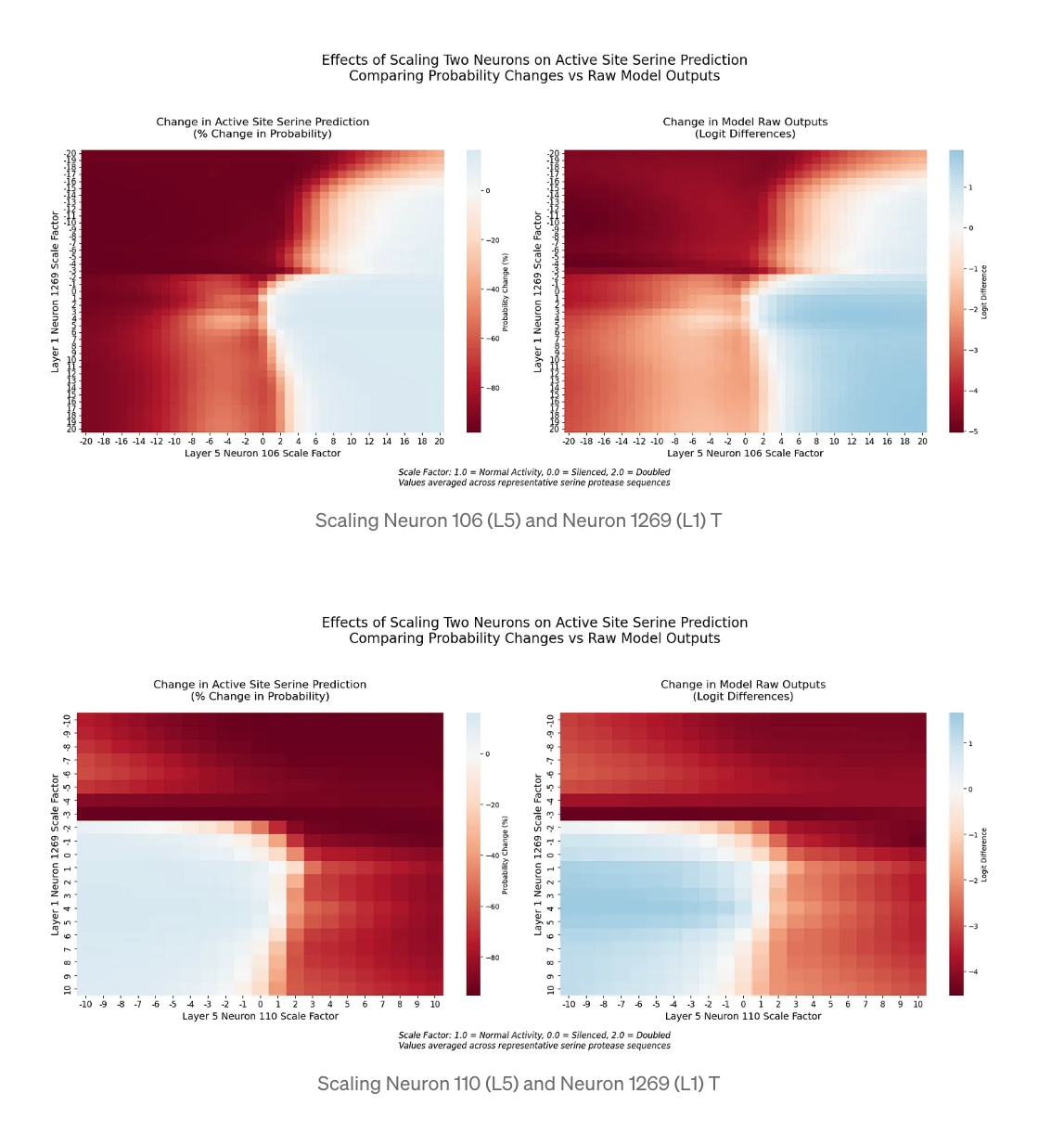
Figure 3: Cross-layer Interaction Effects on Serine Prediction
Heat maps showing the combined effects of scaling neurons across different layers (Layer 1 Neuron 1269 with Layer 5 Neurons 106 and 110) on catalytic serine prediction probability.
Implications
Our findings suggest ESM-2 employs different encoding strategies at different processing depths:
- Early layers contain precise feature detectors
- Later layers integrate this information within broader sequence context
This multi-layer encoding strategy mirrors findings in text-based language models but with layer organization tailored to protein sequences. Finding such clear, compact representations of important biological features demonstrates that mechanistic interpretability techniques can transfer effectively to biological domains.