Breaking Open the Black Box: Making Protein Structure Prediction Interpretable
Nithin Parsan
Co-Founder
Breaking Open the Black Box: Making Protein Structure Prediction Interpretable
December 2024
The Challenge of AI Transparency in Biology
Advances in protein language models like ESMFold and AlphaFold have revolutionized how we predict protein structures from amino acid sequences. These models achieve remarkable accuracy, often rivaling experimental methods. But there's a critical problem: we don't fully understand how they work.
This interpretability gap isn't just an academic concern. Better understanding of these models could enable more targeted protein design, accelerate therapeutic development, and potentially uncover new biological principles.
At Reticular, we've been tackling this challenge head-on, and we're excited to share our latest breakthrough in making protein structure prediction more interpretable.
Our Breakthrough: Seeing Inside Protein Structure Prediction
Our research team has made two key advances:
- Scaling sparse autoencoders to ESM2-3B - We've successfully applied sparse autoencoders to ESM2-3B, the base model for ESMFold, enabling mechanistic interpretability of protein structure prediction for the first time
- Adapting Matryoshka SAEs for protein language models - We've implemented hierarchically organized features by forcing nested groups of latents to reconstruct inputs independently
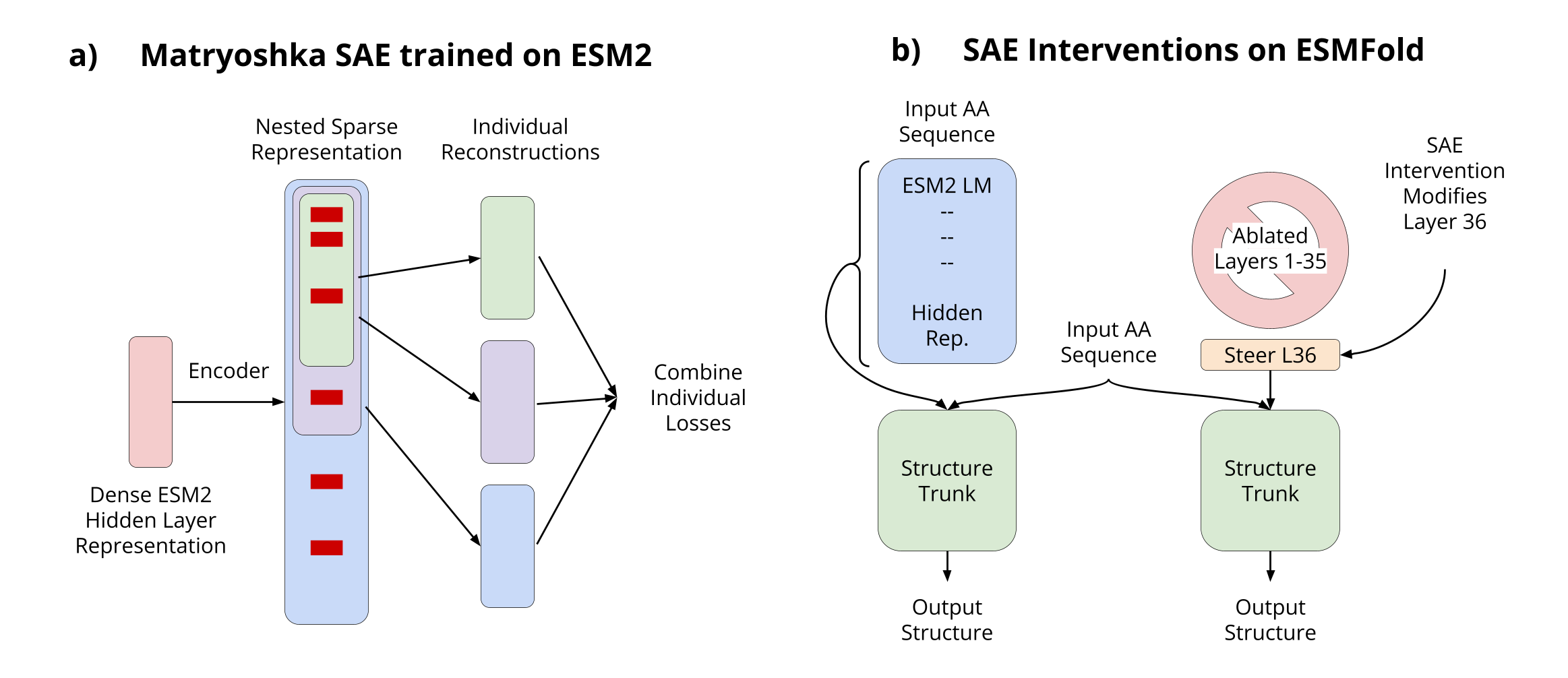
Overview of our approach to interpretable protein structure prediction
This approach allows us to peer inside the "black box" of protein structure prediction models, revealing how they translate sequence information into structural predictions.
Why This Matters for Antibody Engineering
In our work with antibody engineering partners, we've consistently encountered a fundamental challenge: data scarcity makes steering biological AI systems difficult. When developing therapeutic antibodies, validation data is often limited and expensive to generate.
Our sparse autoencoder approach directly addresses this challenge by:
- Enabling precise model steering with minimal data - By understanding exactly how models translate sequence to structure, we can make targeted interventions that require fewer experimental validations
- Providing mechanistic explanations, not just predictions - Our approach explains why certain sequence changes affect structure and function, not just what changes to try
- Building interpretability directly into model capabilities - Rather than treating interpretability as an afterthought, our approach makes it foundational to the modeling process
Key Findings and Implications
Our research yielded several insights that directly impact antibody engineering:
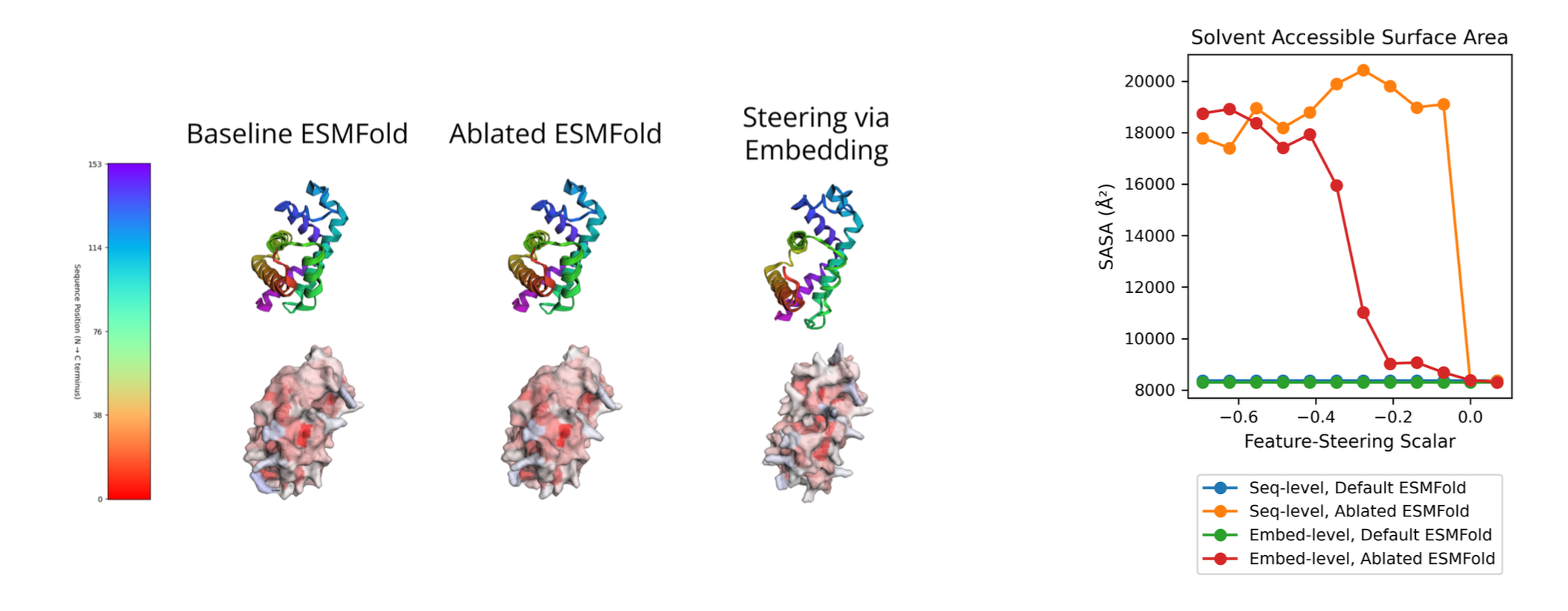
Feature steering demonstration on protein solvent accessibility
- Scale matters significantly for feature interpretability - The jump from 8M to 3B parameters led to a dramatic improvement in biological concept coverage (from ~15-20% to ~49% of concepts)
- Feature steering can control structural properties - We demonstrated the ability to control protein solvent accessibility while maintaining sequence integrity, a capability directly applicable to antibody engineering
- Structure prediction requires surprisingly few features - With only 8-32 active latents per token, SAEs can reasonably recover structure prediction performance, suggesting more efficient modeling approaches are possible
- Hierarchical feature organization aligns with the multi-scale nature of proteins - Our Matryoshka SAE architecture provides a more structured representation that matches biological reality
Interactive Tools for Model Exploration
Beyond the research itself, we've developed a web-based tool that visualizes the hierarchical features discovered by our Matryoshka SAEs. This interface displays how features from different groups activate on protein sequences, connecting sequence patterns directly to structural outcomes.
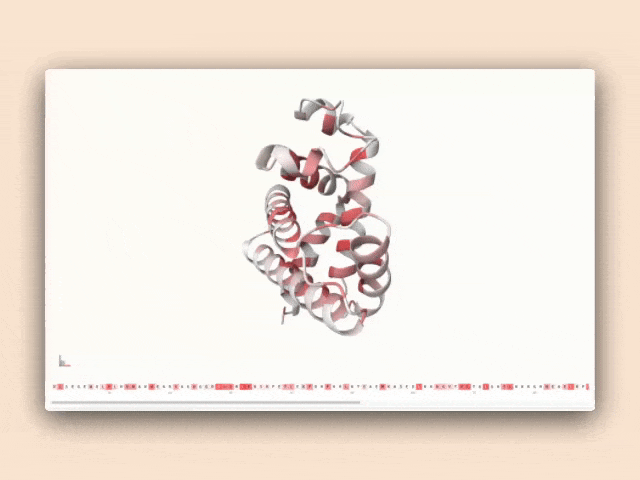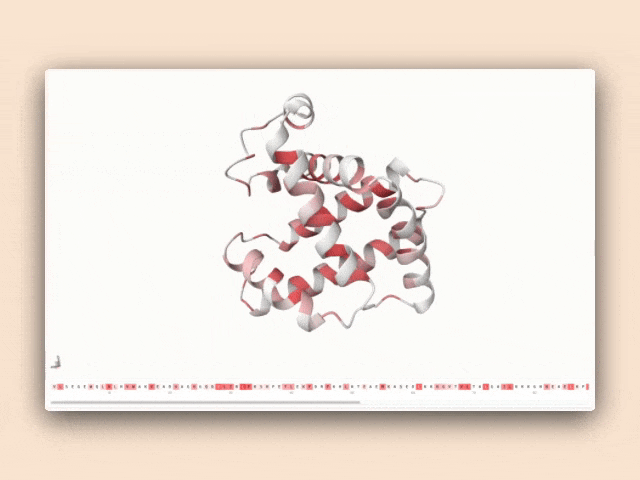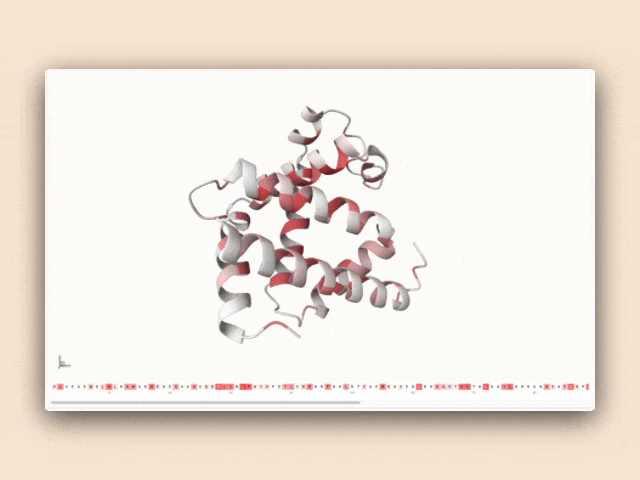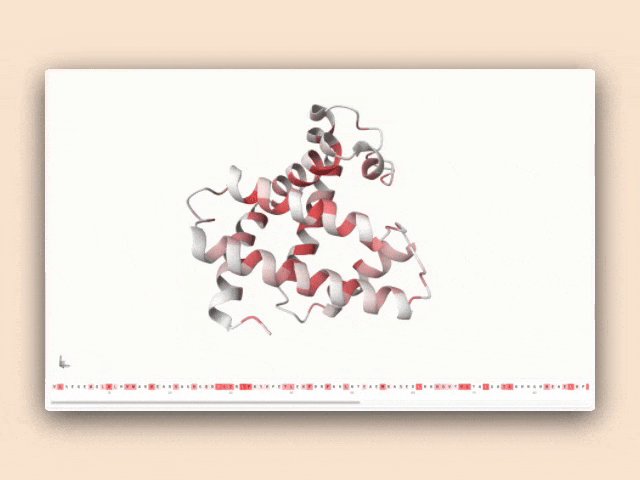
Interactive visualization of protein features
This tool isn't just a demonstration—it's a practical resource for antibody engineers to understand how specific sequence patterns influence structural predictions, bridging the gap between AI research and practical protein engineering applications.
From Research to Real-World Impact
While this research represents a significant technical achievement, its real value lies in practical applications. At Reticular, we're already applying these interpretability techniques to antibody engineering challenges, where they're helping to:
- Reduce experimental iterations by providing mechanistic insights that guide design decisions
- Identify key sequence motifs influencing antibody properties like binding affinity and specificity
- Accelerate development timelines through more targeted and efficient engineering approaches
By providing not just what to try next, but why, we help scientists design smarter experiments with greater confidence.
The Path Forward
This work opens new possibilities for understanding how protein language models translate sequence information into structural predictions, enabling more principled approaches to protein design and engineering.
The code and trained models are available at our GitHub repository, and visualization tools are available at sae.reticular.ai to facilitate further investigation by the research community.
Partner With Us
Are you working on therapeutic antibody development with challenging targets? Our interpretable AI approach can help you accelerate your pipeline, reduce experimental costs, and gain deeper insights into antibody structure-function relationships.
We're currently seeking partnerships with:
- Pharma teams developing therapeutic antibodies with challenging targets
- Biotech companies working with antibody language models
- Research teams exploring mechanistic interpretability for biologics
Transform Your Antibody Engineering Pipeline
Discover how our interpretable AI approach can provide new insights into your antibody candidates and accelerate your development timeline.
Schedule a Demo →Limited early access spots available
Citation: Parsan, N., Yang, D.J., & Yang, J.J. (2025). Towards Interpretable Protein Structure Prediction with Sparse Autoencoders. ICLR GEM Bio Workshop 2025.